Swimming bacteria in flow
The natural habitats of swimming microorganisms, from algae in the oceans to bacteria living in our intestines, are characterized by dynamic fluid flows. The importance of these swimming cells to marine oxygen production, biofilm formation, and the remineralization of organic matter are widely recognized by researchers. However, the interplay of cell motility — the ability of a cell to move independently using metabolic energy — with fluid flow and the porous microstructure of their environments, such as tissues and soils, eludes predictive models of these processes. Few studies in this emerging field have attempted to untangle these complex microscopic interactions, which can lead to macroscopic implications for disease transmission and bioremediation.
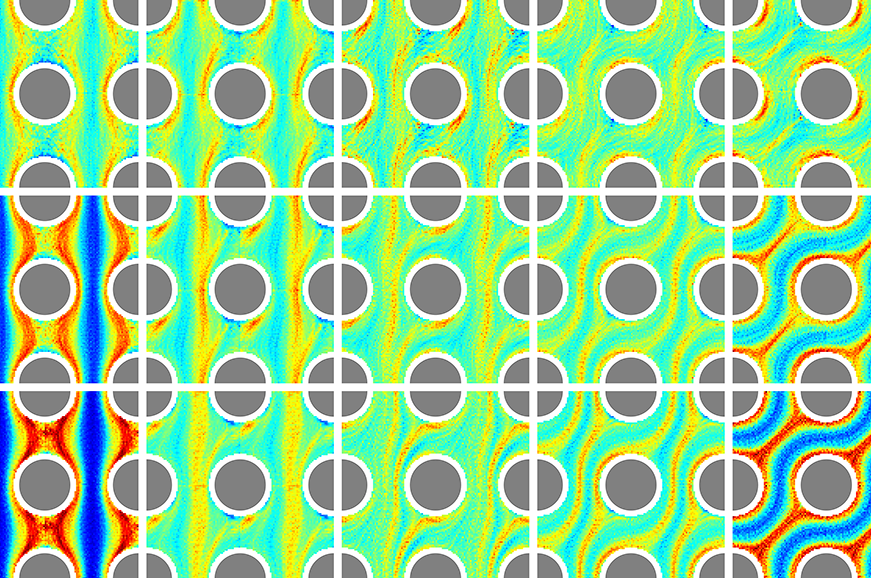
Researchers at Tufts University and the Massachusetts Institute of Technology set out to understand the key physical mechanisms governing the dispersal of swimming microorganisms in porous media flows. The inspiration for this study came from an unlikely source: light scattering in crystalline materials. The team used microfluidic experiments coupled with mathematical simulations to measure how swimming bacteria (Bacillus subtilis) are scattered by flow through a microscopic lattice of obstacles.
In research published in Proceedings of the National Academy of Sciences, the team found that bacteria locally concentrated themselves into filamentous cell density patterns that meandered through the medium and around obstacles. Similar to classical light scattering experiments, varying the flow angle relative to the obstacle lattice caused drastic changes in the filament topologies and the strength of the accumulations. The researchers showed that these filamentous bacterial density patterns closely mimic the density patterns found in the natural habitats of swimming microorganisms.
Researchers discovered that this densification was being driven by the elongated shape of the bacterial cells, which promotes alignment to specific regions of the fluid flow — namely, regions of strong extension that occur near the obstacles in the microfluidic lattice. Once the bacteria are aligned to the flow, fluid advection sweeps the cells downstream promoting the filamentous shape of the cell aggregations.
The researchers observed that bacterial density patterns do not occur at random, but rather their presence is strongly correlated with features of the underlying fluid flow. These density patterns describe regions of the flow that experienced strong stretching. The research team’s new findings may pave the way for new tools to predict where and how swimming microbes accumulate in complex flows, ranging from porous media to marine turbulence. Such tools are key to modeling biogeochemical processes that shape our environment and natural ecosystems.
The study ultimately found that fluid flow gradients and the porous microstructure can be optimized to induce concentrated, directional bacterial dispersion in complex flow environments. Interactions with flow enhanced the spreading of actively swimming bacteria in the flow direction up to 100-fold beyond spreading bacteria without flow. This discovery may provide important new insights into microbial ecosystems, biomedical device design, and the guidance of swimming micro-robots.
The study was conducted by the research team of Ph.D. candidate Amin Dehkharghani, postdoctoral scholar Nicolas Waisbord, and Assistant Professor Jeffrey Guasto of the Tufts Department of Mechanical Engineering, with Associate Professor Jörn Dunkel of the MIT Department of Mathematics. This work was funded through the National Science Foundation.
Department:
Mechanical Engineering